Unravelling CRISPR In The Café
A sketch at a café meeting sets Jennifer Doudna on the path to developing one of the most consequential gene editing tools.
The following is an excerpt from A Crack in Creation: Gene Editing and the Unthinkable Power to Control Evolution, by Jennifer Doudna. Listen to her interview.
I will never forget the first time I heard the term CRISPR.
It was 2006, and I was sitting in my office on the seventh floor of Stanley Hall at the University of California, Berkeley, when the phone rang. On the line was Jillian Banfield, a fellow Berkeley professor from the Departments of Earth and Planetary Science and Environmental Science, Policy, and Management.
I knew Jill only by reputation, and she knew equally little about me; she explained that she had found my lab’s website only after a quick Google search. A geomicrobiologist who focused primarily on the interactions between microorganisms and their environments, Jill had been looking for faculty members at Berkeley who were researching RNA interference, a molecular system that plant and animal cells use to suppress the expression of particular genes and that organisms also use during their immune responses. It was a subject with which my lab had extensive experience.
Jill told me that her lab was studying something I heard as crisper — she didn’t define or even spell the term, only mentioned that it had popped up in some data sets her lab was analyzing — and that she wanted to expand her investigation using tools from genetics and biochemistry, two skill sets my lab could offer. In particular, she thought there might be some parallels between “crisper” and RNA interference. Would I like to meet and discuss it?
[Just how easy is it to edit DNA?]
I was intrigued by Jill’s intensity, although I had my doubts about her request; I had no idea what she was studying, after all. But her excitement was palpable even over the phone, so I agreed to meet her for coffee the following week.
After the call, I did a quick search in the scientific literature and found only a handful of articles on the subject Jill had been so excited about. By comparison, RNA interference, the study of which was scarcely eight years old, had already racked up over four thousand references. (The attention would reach a climax when its discoverers, Andrew Fire and Craig Mello, won a Nobel Prize later that year.) The relative dearth of publications on Jill’s topic made it challenging to evaluate — but it also piqued my curiosity.
I skimmed several of the background articles, reading just enough to learn that this thing — CRISPR — referred to a region of bacterial DNA and that the acronym stood for “clustered regularly interspaced short palindromic repeats.” I didn’t read any further, stymied by the jargon and figuring that Jill would fill me in when we met.
My own Google search showed just how accomplished a scientist Jill was. Brilliant and creatively engaged in multiple diverse areas of science, she’d published articles with titles such as “Mineralogical Biosignatures and the Search for Life on Mars” and “Geophysical Imaging of Stimulated Microbial Biomineralization.” Her research involved collecting and studying samples from far-flung sources, like biospheres deep in the earth beneath Japan, hypersaline lakes in Australia, and acidic mine drainages in Northern California. These exotic projects stood in marked contrast to my own; aside from requiring frequent trips to the x-ray-generating particle accelerator at the Lawrence Berkeley National Laboratory, my lab’s work took place mostly inside test tubes.

Partly because I was so impressed with her research and partly for my own scientific reasons, I was growing keener to meet Jill. I had relocated to Berkeley from Yale University four years earlier with my now-husband, Jamie Cate, and our newborn son, Andrew. Although my ongoing research had moved in some new directions, I was hoping to expand the lab and pick up some additional projects while also cultivating partnerships with new colleagues. This could be just the lead I was looking for.
Jill and I met the following week at the Free Speech Movement Café, near the entrance to one of the campus’s undergraduate libraries. It was a blustery spring day, and when I arrived, Jill was already seated at a stone table in the outdoor courtyard, a notepad and a stack of papers beside her. After we had chatted for a bit, she picked up her notebook and got down to business.
[CRISPR gets a “yellow light” for gene editing.]
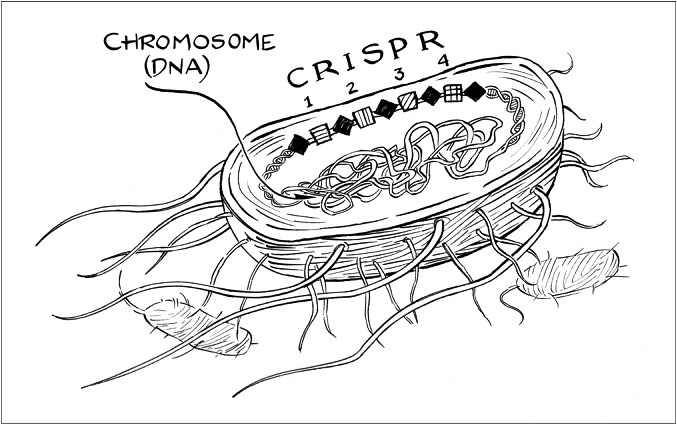
She quickly sketched a diagram of CRISPR. First, she drew a large oval to represent a bacterial cell. Then she drew a circle inside the oval, the bacterial chromosome, and added a series of alternating diamonds and squares along one side of the circle to represent a region of DNA. This region, apparently, was CRISPR.
Jill shaded in the diamonds and mentioned that they were all identical stretches of the same thirty-ish letters of DNA. Then she numbered the squares sequentially beginning at 1 while explaining that each one constituted a unique sequence of DNA.
Finally, the words that the acronym stood for — clustered regularly interspaced short palindromic repeats — began to make sense to me. The diamonds were the short repeats, the squares were interspacing sequences that regularly interrupted the repeats, and these diamond-square arrays were clustered in just one region of the chromosome, not randomly distributed throughout. (When I inspected the repeating DNA sequences more closely, back in my office, the P in the acronym also became clear: the sequences were nearly the same when read in either direction, just like a palindrome such as “senile felines.”)
The basic idea that cells could contain repeating DNA sequences was not itself a surprise; more than 50 percent of the human genome — well over one billion letters of DNA — comprises different types of repetitive arrays, some of which are copied millions of times. Although comparatively smaller bacterial genomes contain far less, I was aware that they too had repeating sequences, some of which shared terms with CRISPR, like repetitive extragenic palindromic (REP) sequences and bacterial interspersed mosaic elements (BIME). But I had never heard of DNA repeating itself with this kind of precision and uniformity, where every repetition was truly identical and always separated from its neighbor by a similarly sized, random spacer sequence.
Curious to learn more about these strange regions of bacterial DNA, I asked Jill about their biological function and was disappointed to hear that she didn’t know. But her lab had uncovered an important clue: DNA sequences from natural bacterial populations showed that essentially every cell had a different CRISPR array due to the unique sequences inter-spaced between the repeats. This was completely unprecedented because every other part of the DNA was nearly identical in each of these cells. Jill recognized that CRISPRs were probably the fastest-evolving regions in the genome, consistent with a function that had to change or adapt quickly in response to something the cells encountered in their environment.
[The history of genetics is a history of ethical conundrums.]
Years earlier, pioneering work by Francisco Mojica, a professor in Spain, had uncovered the same kinds of repeats in numerous completely unrelated species, including in archaea, single-celled microorganisms that, like bacteria, lack nuclei. (Bacteria, Archaea — referred to collectively as prokaryotes — and Eukarya constitute the three domains that encompass all life on earth.) CRISPRs, Jill said, had been found in almost half of all the bacterial genomes that had been sequenced to date and in nearly every archaeal genome. In fact, they appeared to be the most widely shared family of repeating DNA sequences in all prokaryotes.
These bits of information sent a little shiver of intrigue down my spine; if CRISPR was present in so many different species, there was a good chance that nature was using it to do something important.
Excerpted from A Crack in Creation: Gene Editing and the Unthinkable Power to Control Evolution by Jennifer Doudna and Samuel Sternberg. Copyright © 2017 by Jennifer Doudna and Samuel Sternberg. Used by permission of Houghton Mifflin Harcourt Publishing Company. All rights reserved.
Jennifer Doudna is co-author of A Crack in Creation: Gene Editing and the Unthinkable Power to Control Evolution (Houghton Mifflin Harcourt, 2017). She’s an investigator with the Howard Hughes Medical Institute and a professor of Chemistry and Molecular and Cell Biology at the University of California, Berkeley.